Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Collaborative Laboratories for Advanced Decommissioning Science; Keio University*
JAEA-Review 2021-048, 181 Pages, 2022/01
The Collaborative Laboratories for Advanced Decommissioning Science (CLADS), Japan Atomic Energy Agency (JAEA), had been conducting the Nuclear Energy Science & Technology and Human Resource Development Project (hereafter referred to "the Project") in FY2020. The Project aims to contribute to solving problems in the nuclear energy field represented by the decommissioning of the Fukushima Daiichi Nuclear Power Station (1F), Tokyo Electric Power Company Holdings, Inc. (TEPCO). For this purpose, intelligence was collected from all over the world, and basic research and human resource development were promoted by closely integrating/collaborating knowledge and experiences in various fields beyond the barrier of conventional organizations and research fields. The sponsor of the Project was moved from the Ministry of Education, Culture, Sports, Science and Technology to JAEA since the newly adopted proposals in FY2018. On this occasion, JAEA constructed a new research system where JAEA-academia collaboration is reinforced and medium-to-long term research/development and human resource development contributing to the decommissioning are stably and consecutively implemented. Among the adopted proposals in FY2019, this report summarizes the research results of the "Study of corrosion and degradation of the objects in the nuclear reactor by microorganisms" conducted in FY2019 and FY2020. Since the final year of this proposal was FY2020, the results for two fiscal years were summarized. The purpose of the study is to obtain knowledge related to microorganisms that will be useful in the decommissioning process of 1F. Therefore, we clarified the current conditions of the microbial community inhabiting the power plant and its premises. Environmental samples were taken from several sites such as, topsoil from the south of the plant site boundary (south of the treated water tanks), seabed soil and its above water near the plant, surface water 3km offshore …
Collaborative Laboratories for Advanced Decommissioning Science; Keio University*
JAEA-Review 2020-047, 63 Pages, 2021/01
The Collaborative Laboratories for Advanced Decommissioning Science (CLADS), Japan Atomic Energy Agency (JAEA), had been conducting the Nuclear Energy Science & Technology and Human Resource Development Project (hereafter referred to "the Project") in FY2019. The Project aims to contribute to solving problems in the nuclear energy field represented by the decommissioning of the Fukushima Daiichi Nuclear Power Station, Tokyo Electric Power Company Holdings, Inc. (TEPCO). For this purpose, intelligence was collected from all over the world, and basic research and human resource development were promoted by closely integrating/collaborating knowledge and experiences in various fields beyond the barrier of conventional organizations and research fields. The sponsor of the Project was moved from the Ministry of Education, Culture, Sports, Science and Technology to JAEA since the newly adopted proposals in FY2018. On this occasion, JAEA constructed a new research system where JAEA-academia collaboration is reinforced and medium-to-long term research/development and human resource development contributing to the decommissioning are stably and consecutively implemented. Among the adopted proposals in FY2019, this report summarizes the research results of the "Study of corrosion and degradation of the objects in the nuclear reactor by microorganisms" conducted in FY2019. The purpose of the study is to obtain knowledge related to microorganisms that will be useful in the decommissioning process of the Fukushima Daiichi Nuclear Power Station. For this reason, the current conditions of the microbial community inhabiting the power plant and its premises will be clarified. In the first research year, we obtained environmental samples such as soils from the south of the boundary of the plant, seabed soils near the plant, and surface water 3 km offshore from the plant, and successfully prepared their microbial genomic DNAs.
Balestrazzi, A.*; Achary V Mohan Murali*; Macovei, A.*; Yoshiyama, Kaoru*; Sakamoto, Ayako
Frontiers in Plant Science (Internet), 7, p.64_1 - 64_2, 2016/02
Times Cited Count:3 Percentile:56.08(Plant Sciences)no abstracts in English
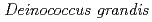 , isolated from freshwater fish in Japan
, isolated from freshwater fish in JapanSato, Katsuya; Onodera, Takefumi*; Omoso, Kota*; Takeda-Yano, Kiyoko*; Katayama, Takeshi*; Ono, Yutaka; Narumi, Issey*
Genome Announcements (Internet), 4(1), p.e01631-15_1 - e01631-15_2, 2016/01
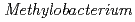 sp. ME121, isolated from soil as a mixed single colony with
sp. ME121, isolated from soil as a mixed single colony with 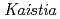 sp. 32K
sp. 32KFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Shimizu, Tetsu*; Wakabayashi, Yu*; Narumi, Issey*; Nakamura, Akira*; Ito, Masahiro*
Genome Announcements (Internet), 3(5), p.e01005-15_1 - e01005-15_2, 2015/09
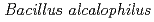 AV1934, a classic alkaliphile isolated from human feces in 1934
AV1934, a classic alkaliphile isolated from human feces in 1934Attie, O.*; Jayaprakash, A.*; Shah, H.*; Paulsen, I. T.*; Morino, Masato*; Takahashi, Yuka*; Narumi, Issey*; Sachidanandam, R.*; Sato, Katsuya; Ito, Masahiro*; et al.
Genome Announcements (Internet), 2(6), p.e01175-14_1 - e01175-14_2, 2014/11
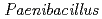 sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ions
sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ionsFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Takahashi, Yuka*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 2(5), p.e00866-14_1 - e00866-14_2, 2014/09
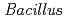 sp. strain TS-2, isolated from a jumping spider
sp. strain TS-2, isolated from a jumping spiderFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 2(3), p.e00458-14_1 - e00458-14_2, 2014/05
Yamaguchi, Akihiro*; Iwadate, Mitsuo*; Suzuki, Eiichiro*; Yura, Kei; Kawakita, Shigetsune*; Umeyama, Hideaki*; Go, Michiko*
Nucleic Acids Research, 31(1), p.463 - 468, 2003/01
Times Cited Count:15 Percentile:25.19(Biochemistry & Molecular Biology)Enlarged FAMSBASE is a relational database of comparative protein structure models for the whole genome of 41 species, presented in the GTOP database. The models are calculated by FAMS, Full Automatic Modeling System. Enlarged FAMSBASE provides a wide range of query keys, such as name of ORF (open reading frame), ORF keywords, PDB ID, PDB heterogen atoms, and sequence similarity. Heterogen atoms in PDB include cofactors, ligands, and other factors that interact with proteins, and are a good starting point for analyzing interactions between proteins and other molecules. The data may also work as a template for drug design. The present number of ORFs with protein 3D models in FAMSBASE is 183,805, and the database includes an average of three models for each ORF. FAMSBASE is available at http://famsbase.bio.nagoya-u.ac.jp/famsbase/.
; Hotta, Hiroshi
Radiat.Res., 64(3), p.407 - 415, 1975/03
Times Cited Count:3no abstracts in English